I am an AI/machine learning and data science practitioner who is passionate about tackling challenging problems in applied areas.
In my professional career, I apply machine learning, deep learning, and statistical inference to challenges in student latent state modeling and educational content recommendation. I also work with large language models and generative AI models to develop innovative tools for enriching curriculum and assessment. Previously, I worked on projects related to scientific instrumentation, including cell image analysis with deep learning and flow cytometry data analysis and collection. I also have professional experience in traditional software engineering and data analysis. My academic background is in computer science, statistics, and bioinformatics. In my graduate research, I focused primarily on machine learning, deep learning, and statistics with applications in tissue engineering.
Research
A selection of my academic and professional research.
LENS: Uncertainty-preserving deep knowledge tracing with state-space models
S. Thomas Christie, Carson Cook, Anna Rafferty. Published: Proceedings of the 17th International Conference on Educational Data Mining (2024).
A central goal of both knowledge tracing and traditional assessment is to quantify student knowledge and skills at a given point in time. Deep knowledge tracing flexibly considers a student's response history but does not quantify epistemic uncertainty, while IRT and CDM compute measurement error but only consider responses to individual tests in isolation from a student's past responses. Elo and BKT could bridge this divide, but the simplicity of the underlying models limits information sharing across skills and imposes strong inductive biases. To overcome these limitations, we introduce Dynamic LENS, a modeling paradigm that combines the flexible uncertainty-preserving properties of variational autoencoders with the principled information integration of Bayesian state-space models.
Dynamic LENS allows information from student responses to be collected across time, while treating responses from the same test as exchangeable observations generated by a shared latent state. It represents student knowledge as Gaussian distributions in high-dimensional space and combines estimates both within tests and across time using Bayesian updating. We show that Dynamic LENS has similar predictive performance to competing models, while preserving the epistemic uncertainty - the deep learning analogue to measurement error - that DKT models lack. This approach provides a conceptual bridge across an important divide between models designed for formative practice and summative assessment.
LENS: Predictive Diagnostics for Flexible and Efficient Assessments
S. Thomas Christie, Hayden Johnson, Carson Cook, Garron Gianopulos, Anna Rafferty. Published: Proceedings from L@S (2023).
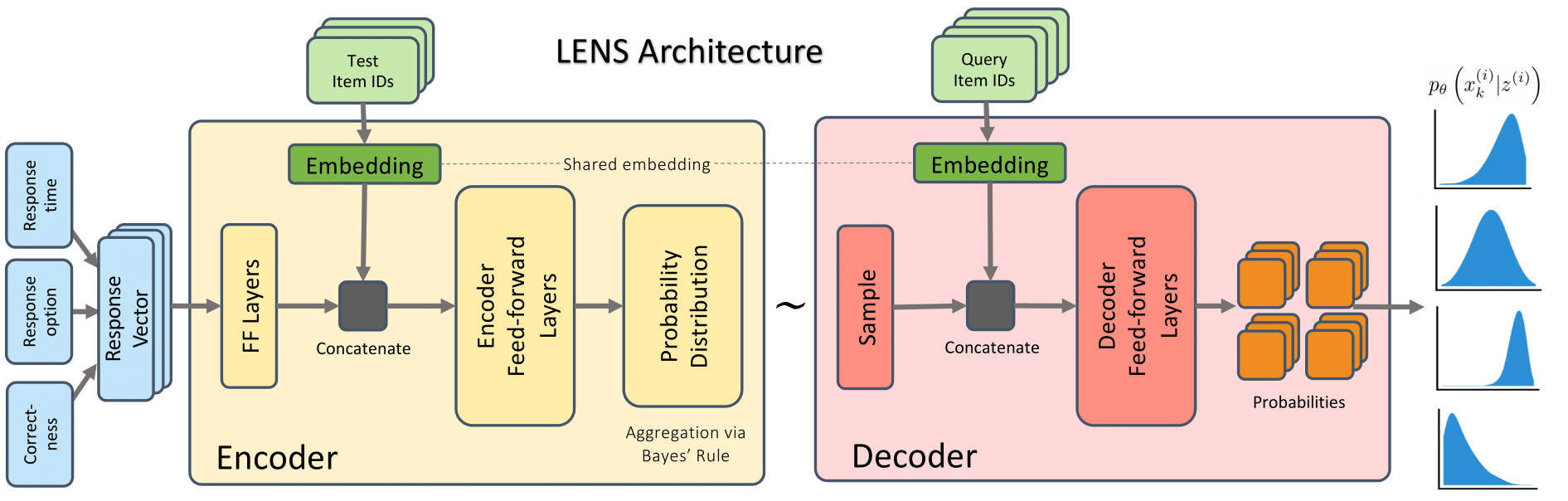
The utility of assessment systems lies in their capacity to transform observations of student behavior into meaningful inferences about learning, knowledge, and skills. Common practice is to use latent variable models and produce scores on scales. However the simplicity of these psychometric models may filter out potentially valuable information present in student behavior. In particular, scale scores are not optimized to support granular instructional decisions. Machine learning offers promising alternatives, but proposed deep learning architectures are not ideally suited for operational testing conditions involving sparse data and shifting category labels for test questions.
We present criteria for a model architecture that can leverage the rich, sparse behavioral data made available by modern assessment systems. We argue that behavioral predictions with well-quantified uncertainties are a viable alternative to scale scores for many applications. To satisfy these goals, we propose the LENS model architecture that combines the flexibility of machine learning and the uncertainty quantification of latent variable models to produce high-quality predictive diagnostic claims. Inspired by variational autoencoders, LENS maps student behavior to a latent probability distribution. In addition, LENS performs explicit Bayesian integration of each behavioral observation, allowing the model to incorporate sparse data with informative prior beliefs about students.
We then compare the predictive capability of LENS to both latent variable and machine learning approaches. LENS generates competitive or superior predictions of behavior, particularly on sparse data, while requiring fewer modeling assumptions than other options and allowing for easy incorporation of auxiliary behavioral data. We also show how to produce interpretable claims about student behavior at multiple levels of granularity, allowing the same model to serve multiple reporting needs. Finally, we discuss the flexibility afforded by the proposed model for constructing assessments customized to both educator needs and student skills.
Student State Space Modeling with Bayesian Information Integration
Carson Cook, Garron Gianopulos, Emily Lubkert, Yeow Meng Thum, and S. Thomas Christie. Presented: NCME Annual Meeting (2023), NCSA (2023).
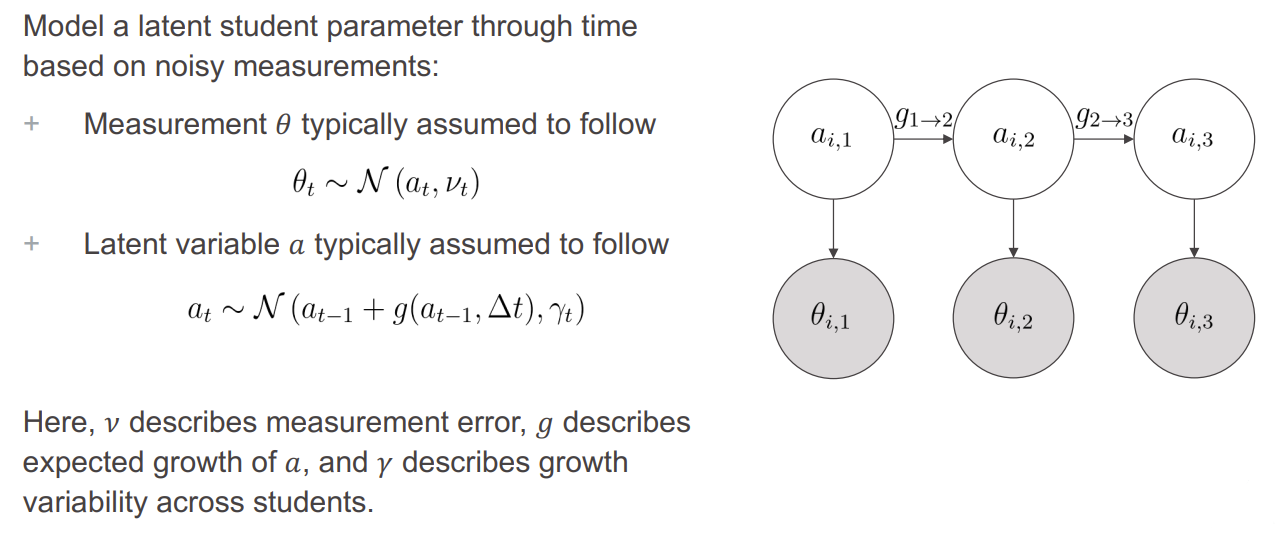
In the United States, summative assessments - standardized tests taken at a single point in time, typically at the end of the school year - are widely used for accountability purposes; however, the results of these assessments come too late in the school year to have an impact on classroom instruction. In response, many districts look to interim assessments, which are administered multiple times through the school year, for more timely feedback on the student progress. Through-year assessments are an emerging alternative where the goal is frequently to administer interim assessments and use their results to arrive at summative scores. This approach is intended to satisfy the twin objectives of (1) providing timely feedback on student progress and (2) eliminate the need for a separate summative test to reduce testing burden. Unfortunately, this creates another challenge: how should multiple test scores be combined to produce a single summative score? If the goal is to arrive at a summative score that combines information across tests, is measurement-centric, and requires few hand-tuning decisions, there is currently no favored method.
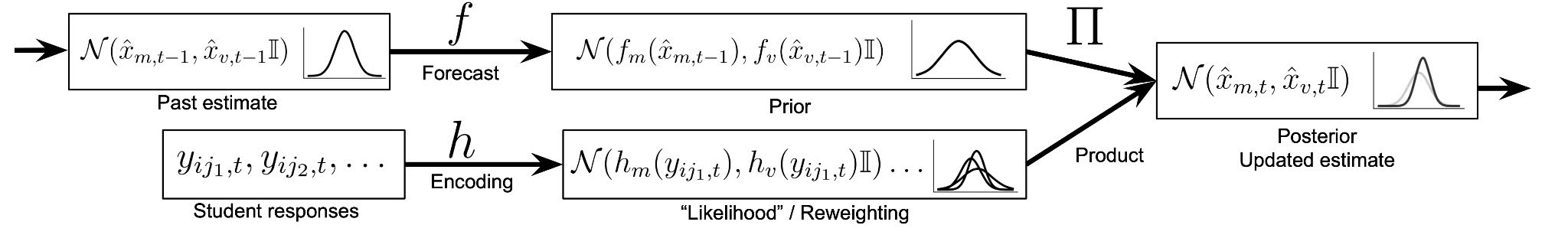
To address this challenge, we created a Bayesian state space model, inspired by the Kalman filter, that combines information across test events to arrive at a composite score that can be used for summative purposes. Our model uses three test scores - fall, winter, and spring - together with expected growth to produce an estimate of student proficiency in the spring. We fit our model to simulated data and found that, though the estimates produced by our model were somewhat biased for simulated students with atypical growth, it performed substantially better in terms of RMSE than the using the spring score alone. We also evaluated the model fit on a historical MAP Growth dataset, via Bayesian model comparison, and found that the proposed model is more likely to have generated the data than the baseline model, which simply uses spring scores as the ability estimates.
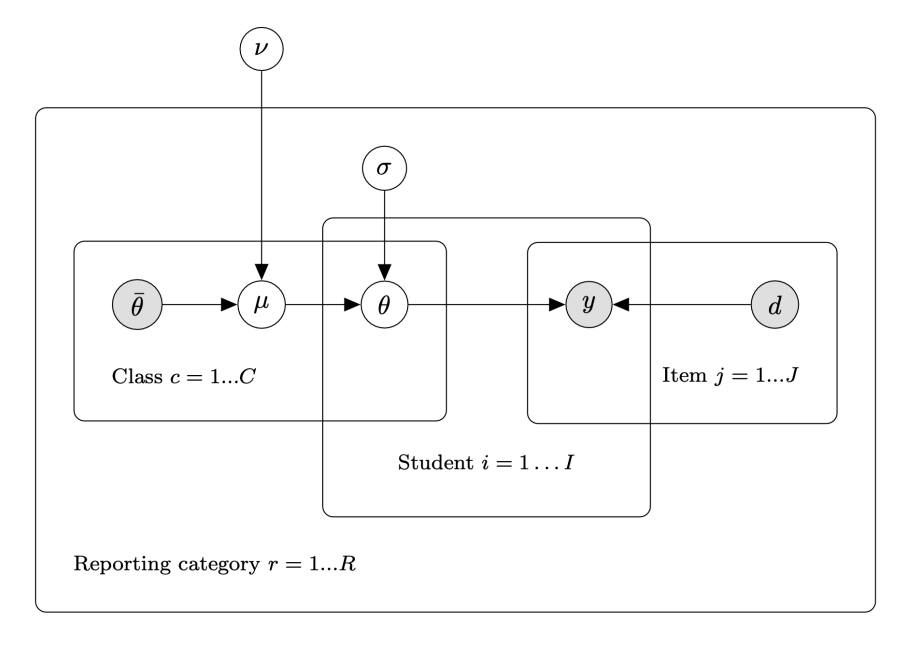
Group-Level Ability Estimation Using Bayesian Multilevel Modeling: Opportunities and Limitations
S. Thomas Christie, Carson Cook, and Garron Gianopulos. Presented: NCME Annual Meeting (2023), NCSA (2023).
Granular, instructionally-relevant sub-scores have the potential to increase the utility of standardized assessment reports for teachers in the classroom. However, existing procedures tend to produce sub-scores with poor reliability due to the small number of item responses used to compute them. Since instructional planning decisions are often made for groups of students, we introduce and evaluate a model for group-level sub-scores that allows us to pool information across students and produces one sub-score per reporting category per student group. Our primary interest is whether coordinate shift in performance by students is detectable given our method. Specifically, we investigate whether a granular class-level score reveals a detectable difference in performance within specific reporting categories when compared to the responses predicted by overall scores. Through this lens, a sub-score would identify class-wide areas of strength or opportunities for class-level instruction, thereby assisting teachers with instructional planning.
To validate the proposed approach, we conducted a simulation study and determined the effect size, class size, and the number of item responses required to detect class-level performance differences within granular item reporting categories. We then applied our modeling approach to historical ELA and Math assessment data to characterize its potential utility in those domains. Our findings indicate that this method may have utility for identifying variation in class-level performance at the level of reporting categories, but this is highly dependent on the test blueprint and the magnitude of the effect.
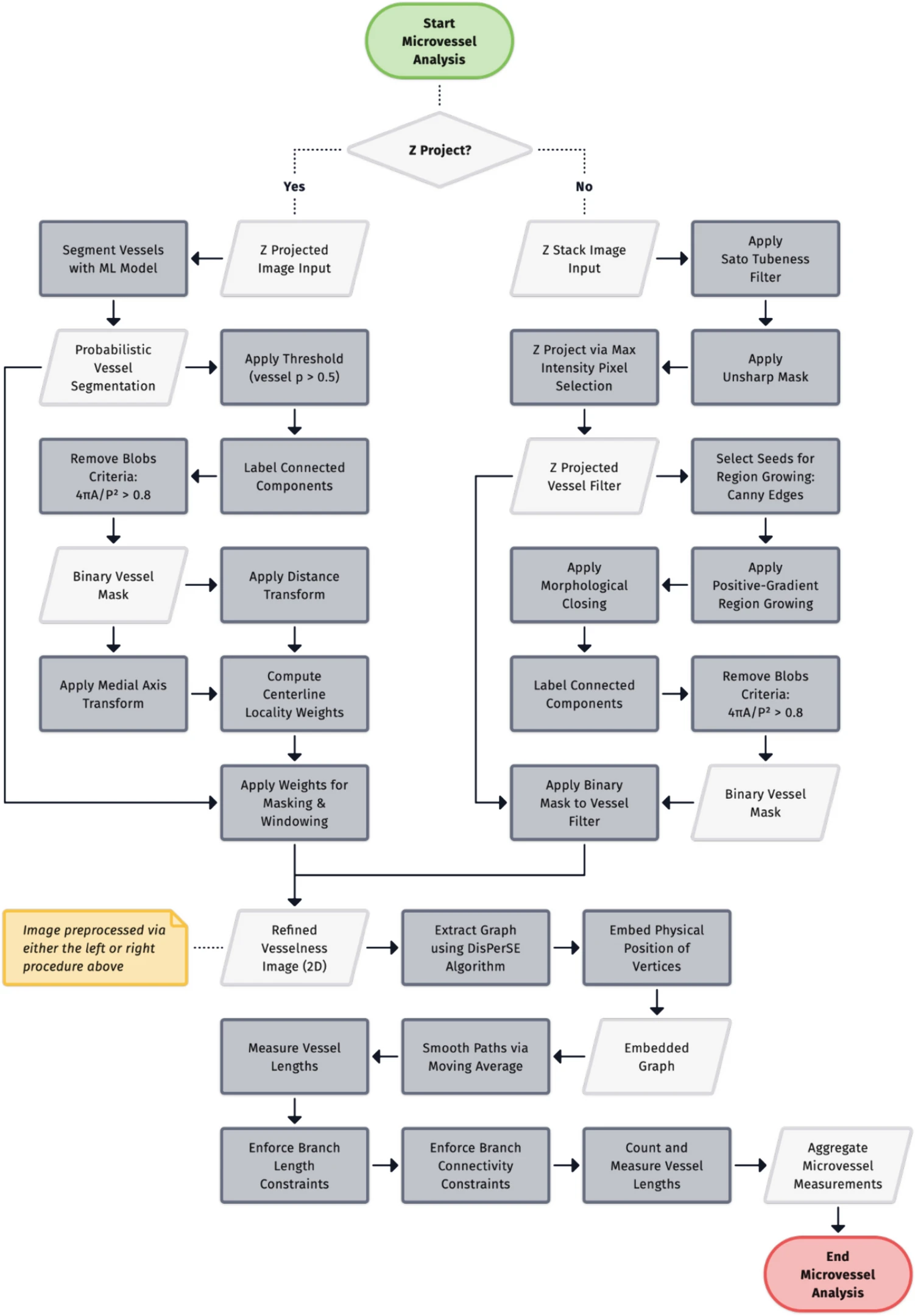
Empowering High-Throughput High-Content Analysis of Microphysiological Models: Open-Source Software for Automated Image Analysis of Microvessel Formation and Cell Invasion
Noah Wiggin, Carson Cook, Mitchell Black, Ines Cadena, Salam Rahal-Arabi, Chandler L. Asnes, Yoanna Ivanova, Marian H Hettiaratchi, Laurel E Hind, Kaitlin C Fogg. Published: Cellular and Molecular Bioengineering (2024).
The primary aim of this study was to develop an open-source Python-based software for the automated analysis of dynamic cell behaviors in microphysiological models using non-confocal microscopy. This research seeks to address the existing gap in accessible tools for high-throughput analysis of endothelial tube formation and cell invasion in vitro, facilitating the rapid assessment of drug sensitivity.
Our approach involved annotating over 1000 2 mm Z-stacks of cancer and endothelial cell co-culture model and training machine learning models to automatically calculate cell coverage, cancer invasion depth, and microvessel dynamics. Specifically, cell coverage area was computed using focus stacking and Gaussian mixture models to generate thresholded Z-projections. Cancer invasion depth was determined using a ResNet-50 binary classification model, identifying which Z-planes contained invaded cells and measuring the total invasion depth. Lastly, microvessel dynamics were assessed through a U-Net Xception-style segmentation model for vessel prediction, the DisPerSE algorithm to extract an embedded graph, then graph analysis to quantify microvessel length and connectivity. To further validate our software, we reanalyzed an image set from a high-throughput drug screen involving a chemotherapy agent on a 3D cervical and endothelial co-culture model. Lastly, we applied this software to two naive image datasets from coculture lumen and microvascular fragment models.
The software accurately measured cell coverage, cancer invasion, and microvessel length, yielding drug sensitivity IC50 values with a 95% confidence level compared to manual calculations. This approach significantly reduced the image processing time from weeks down to hours. Furthermore, the software was able to calculate cell coverage, microvessel length, and invasion depth from two additional microphysiological models that were imaged with confocal microscopy, highlighting the versatility of the software.
Our free and open source software offers an automated solution for quantifying 3D cell behavior in microphysiological models assessed using non-confocal microscopy, providing the broader Cellular and Molecular Bioengineering community with an alternative to standard confocal microscopy paired with proprietary software.
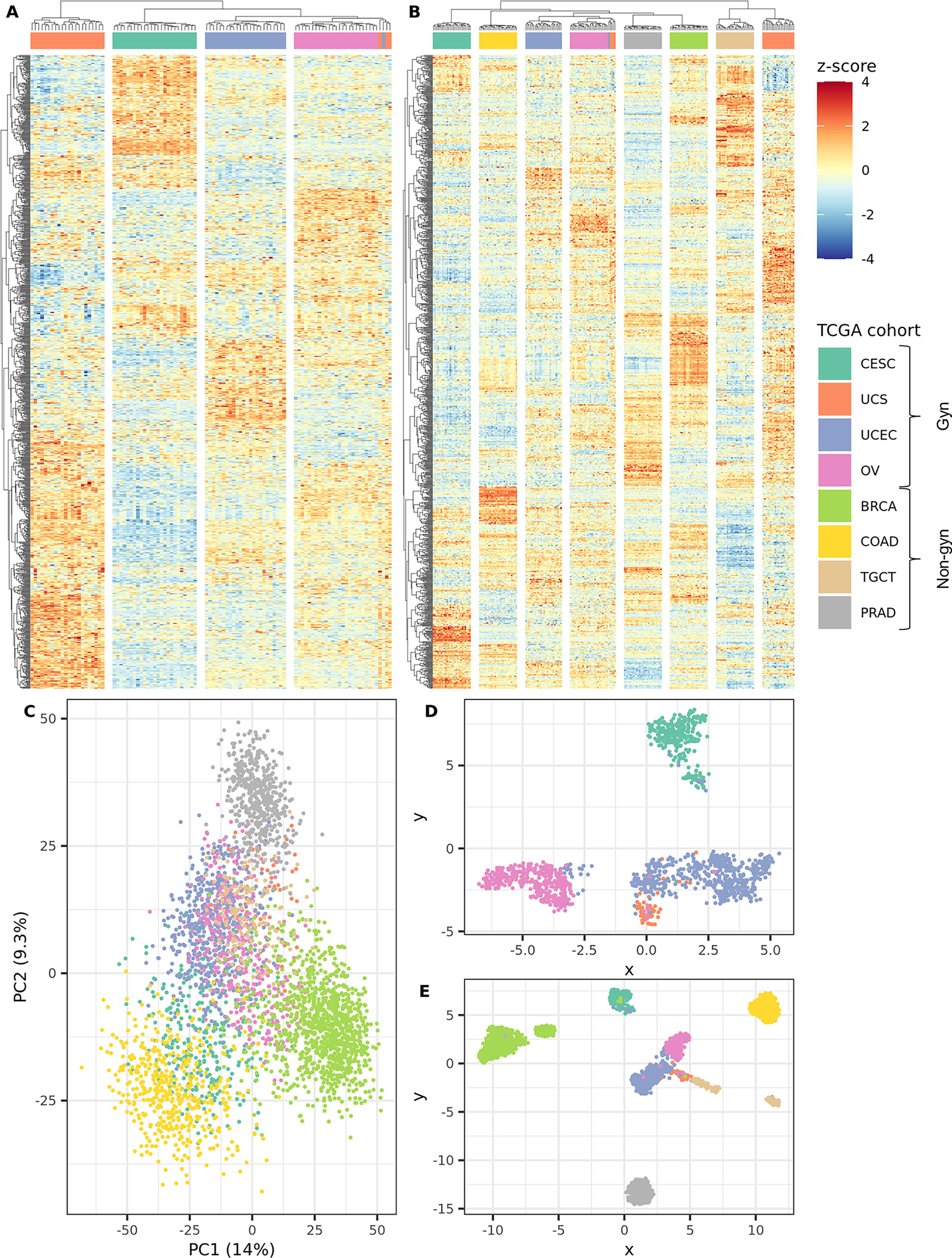
Characterizing the Extracellular Matrix Transcriptome of Endometriosis
Carson Cook, Noah Wiggin, and Kaitlin C. Fogg. Published: Reproductive Sciences (2023).
In recent years, the matrisome, a set of proteins that make up the extracellular matrix (ECM) or are closely involved in ECM behavior, has been shown to have great importance for characterizing and understanding disease pathogenesis and progression. The matrisome is especially critical for examining diseases characterized by extensive tissue remodeling. Endometriosis is characterized by the extrauterine growth of endometrial tissue, making it an ideal condition to study through the lens of matrisome gene expression. While large gene expression datasets have become more available and gene dysregulation in endometriosis has been the target of several studies, the gene expression profile of the matrisome specifically in endometriosis has not been well characterized. In our study, we explored four Gene Expression Omnibus (GEO) DNA microarray datasets containing eutopic endometrium of people with and without endometriosis. After batch correction, menstrual cycle phase accounted for 53% of variance and disease accounted for 23%; thus, the data were separated by menstrual cycle phase before performing differential expression analysis, statistical and machine learning modeling, and enrichment analysis. We established that matrisome gene expression alone can effectively differentiate endometriosis samples from healthy ones, demonstrating the potential of matrisome gene expression for diagnostic applications. Furthermore, we identified specific matrisome genes and gene networks whose expression can distinguish endometriosis stages I/II from III/IV. Taken together, these findings may aid in developing future in vitro models of disease, offer insights into novel treatment strategies, and advance diagnostic tools for this underserved patient population.

Characterizing the extracellular matrix transcriptome of cervical, endometrial, and uterine cancers
Carson Cook, Andrew Miller, Thomas Barker, Yanming Di, Kaitlin Fogg. Published: Matrix Biology Plus (2022).
Increasingly, the matrisome, a set of proteins that form the core of the extracellular matrix (ECM) or are closely associated with it, has been demonstrated to play a key role in tumor progression. However, in the context of gynecological cancers, the matrisome has not been well characterized. A holistic, yet targeted, exploration of the tumor microenvironment is critical for better understanding the progression of gynecological cancers, identifying key biomarkers for cancer progression, establishing the role of gene expression in patient survival, and for assisting in the development of new targeted therapies. In this work, we explored the matrisome gene expression profiles of cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), uterine corpus endometrial carcinoma (UCEC), and uterine carcinosarcoma (UCS) using publicly available RNA-seq data from The Cancer Genome Atlas (TCGA) and The Genotype-Tissue Expression (GTEx) portal. We hypothesized that the matrisomal expression patterns of CESC, UCEC, and UCS would be highly distinct with respect to genes which are differentially expressed and hold inferential significance with respect to tumor progression, patient survival, or both. Through a combination of statistical and machine learning analysis techniques, we identified sets of genes and gene networks which characterized each of the gynecological cancer cohorts. Our findings demonstrate that the matrisome is critical for characterizing gynecological cancers and transcriptomic mechanisms of cancer progression and outcome. Furthermore, while the goal of pan-cancer transcriptional analyses is often to highlight the shared attributes of these cancer types, we demonstrate that they are highly distinct diseases which require separate analysis, modeling, and treatment approaches. In future studies, matrisome genes and gene ontology terms that were identified as holding inferential significance for cancer stage and patient survival can be evaluated as potential drug targets and incorporated into in vitro models of disease.
Personal Projects
A selection of personal projects demonstrating my expertise in machine learning and data science.
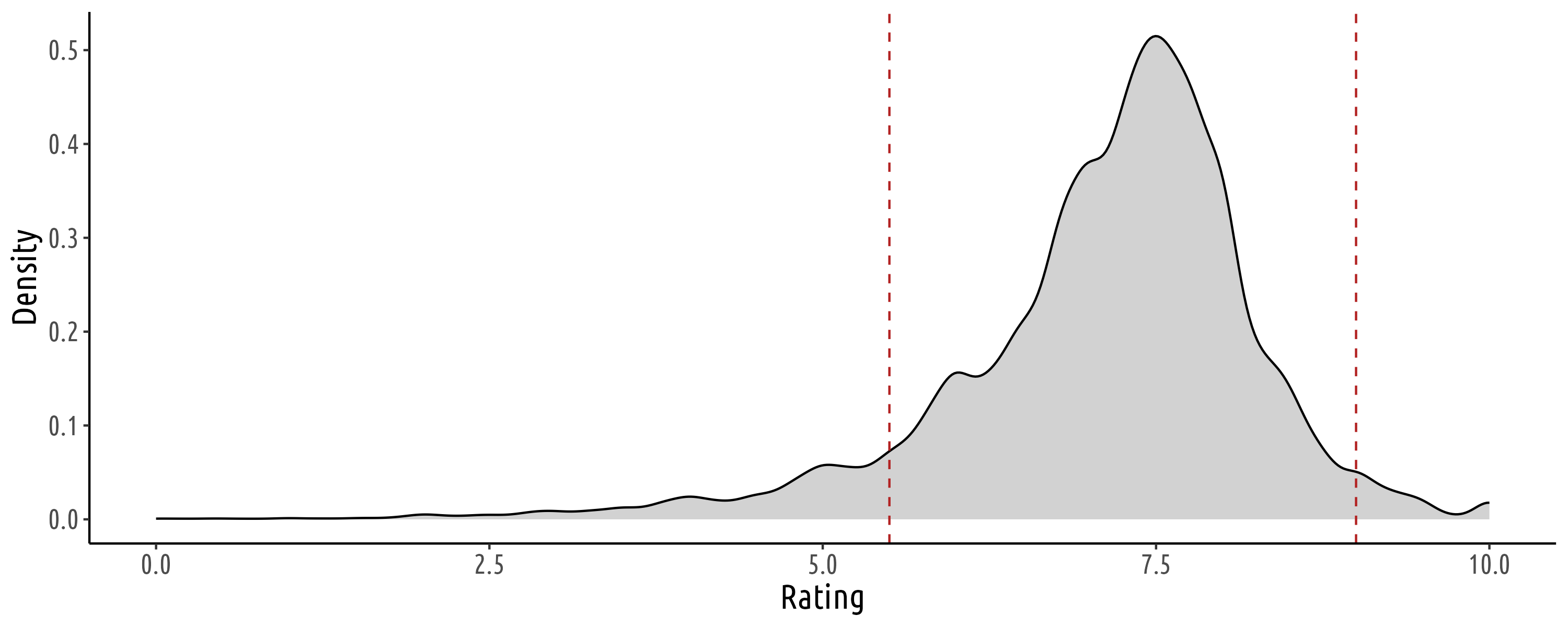
Predicting Pitchfork Album Ratings from Review Text
Goal
This project provides a start-to-finish example of an NLP transfer learning task, detailing data cleaning, exploratory data analysis, model fitting, and post-fit model prediction analysis.
Summary
In this project, I built a regression model to predict album ratings based on review text body. This was accomplished by building a regressor on top of a pre-trained transformer model and performing task-specific fine-tuning. This sequence of notebooks utilizes a Pitchfork reviews dataset of approximately 20K album reviews (mattismegevand/pitchfork). The project consists of a sequence of jupyter notebooks and a corresponding custom library. The notebooks are available as both the original .ipynb source and .html exports.
Links
Education
M.S. Bioengineering (M), Statistics (m), Oregon State University (2022)
Focus on computational transcriptomics and image analysis using statistical inference, machine learning, and deep learning.
Advisors: Kaitlin Fogg (M), Yanming Di (m)
Lab: The Fogg Lab
B.S. Computer Science (M), Mathematics (m), Portland State University (2020)
Focus on machine learning and data science. Participated in MECOP (Multiple Engineering Cooperative Program).